Regularization#
This notebook explores regularization in linear models.
Introductory example#
We demonstrate a common issue with correlated features when fitting linear models.
We use the penguins dataset to illustrate this issue.
# When using JupyterLite, uncomment and install the `skrub` package.
%pip install skrub
import matplotlib.pyplot as plt
import skrub
skrub.patch_display() # makes nice display for pandas tables
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/bin/python: No module named pip
Note: you may need to restart the kernel to use updated packages.
import pandas as pd
penguins = pd.read_csv("../datasets/penguins.csv")
penguins
Processing column 1 / 17
Processing column 2 / 17
Processing column 3 / 17
Processing column 4 / 17
Processing column 5 / 17
Processing column 6 / 17
Processing column 7 / 17
Processing column 8 / 17
Processing column 9 / 17
Processing column 10 / 17
Processing column 11 / 17
Processing column 12 / 17
Processing column 13 / 17
Processing column 14 / 17
Processing column 15 / 17
Processing column 16 / 17
Processing column 17 / 17
| studyName | Sample Number | Species | Region | Island | Stage | Individual ID | Clutch Completion | Date Egg | Culmen Length (mm) | Culmen Depth (mm) | Flipper Length (mm) | Body Mass (g) | Sex | Delta 15 N (o/oo) | Delta 13 C (o/oo) | Comments | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | PAL0708 | 1 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A1 | Yes | 2007-11-11 | 39.1 | 18.7 | 181.0 | 3750.0 | MALE | Not enough blood for isotopes. | ||
| 1 | PAL0708 | 2 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A2 | Yes | 2007-11-11 | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE | 8.94956 | -24.69454 | |
| 2 | PAL0708 | 3 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A1 | Yes | 2007-11-16 | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE | 8.36821 | -25.33302 | |
| 3 | PAL0708 | 4 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A2 | Yes | 2007-11-16 | Adult not sampled. | |||||||
| 4 | PAL0708 | 5 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N3A1 | Yes | 2007-11-16 | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE | 8.76651 | -25.32426 | |
| 339 | PAL0910 | 64 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N98A2 | Yes | 2009-11-19 | 55.8 | 19.8 | 207.0 | 4000.0 | MALE | 9.70465 | -24.53494 | |
| 340 | PAL0910 | 65 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N99A1 | No | 2009-11-21 | 43.5 | 18.1 | 202.0 | 3400.0 | FEMALE | 9.37608 | -24.40753 | Nest never observed with full clutch. |
| 341 | PAL0910 | 66 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N99A2 | No | 2009-11-21 | 49.6 | 18.2 | 193.0 | 3775.0 | MALE | 9.4618 | -24.70615 | Nest never observed with full clutch. |
| 342 | PAL0910 | 67 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N100A1 | Yes | 2009-11-21 | 50.8 | 19.0 | 210.0 | 4100.0 | MALE | 9.98044 | -24.68741 | |
| 343 | PAL0910 | 68 | Chinstrap penguin (Pygoscelis antarctica) | Anvers | Dream | Adult, 1 Egg Stage | N100A2 | Yes | 2009-11-21 | 50.2 | 18.7 | 198.0 | 3775.0 | FEMALE | 9.39305 | -24.25255 |
studyName
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Sample Number
Int64DType- Null values
- 0 (0.0%)
- Unique values
- 152 (44.2%)
- Mean ± Std
- 63.2 ± 40.4
- Median ± IQR
- 58 ± 66
- Min | Max
- 1 | 152
Species
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Region
ObjectDType- Null values
- 0 (0.0%)
Island
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Stage
ObjectDType- Null values
- 0 (0.0%)
Individual ID
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 190 (55.2%)
Most frequent values
Clutch Completion
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (0.6%)
Most frequent values
Date Egg
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 50 (14.5%)
Most frequent values
Culmen Length (mm)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 164 (47.7%)
- Mean ± Std
- 43.9 ± 5.46
- Median ± IQR
- 44.4 ± 9.30
- Min | Max
- 32.1 | 59.6
Culmen Depth (mm)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 80 (23.3%)
- Mean ± Std
- 17.2 ± 1.97
- Median ± IQR
- 17.3 ± 3.10
- Min | Max
- 13.1 | 21.5
Flipper Length (mm)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 55 (16.0%)
- Mean ± Std
- 201. ± 14.1
- Median ± IQR
- 197. ± 23.0
- Min | Max
- 172. | 231.
Body Mass (g)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 94 (27.3%)
- Mean ± Std
- 4.20e+03 ± 802.
- Median ± IQR
- 4.05e+03 ± 1.20e+03
- Min | Max
- 2.70e+03 | 6.30e+03
Sex
ObjectDType- Null values
- 10 (2.9%)
- Unique values
- 3 (0.9%)
Most frequent values
Delta 15 N (o/oo)
Float64DType- Null values
- 14 (4.1%)
- Unique values
- 330 (95.9%)
- Mean ± Std
- 8.73 ± 0.552
- Median ± IQR
- 8.65 ± 0.879
- Min | Max
- 7.63 | 10.0
Delta 13 C (o/oo)
Float64DType- Null values
- 13 (3.8%)
- Unique values
- 331 (96.2%)
- Mean ± Std
- -25.7 ± 0.794
- Median ± IQR
- -25.8 ± 1.27
- Min | Max
- -27.0 | -23.8
Comments
ObjectDType- Null values
- 290 (84.3%)
- Unique values
- 10 (2.9%)
Most frequent values
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | studyName | ObjectDType | 0 (0.0%) | 3 (0.9%) | |||||
| 1 | Sample Number | Int64DType | 0 (0.0%) | 152 (44.2%) | 63.2 | 40.4 | 1 | 58 | 152 |
| 2 | Species | ObjectDType | 0 (0.0%) | 3 (0.9%) | |||||
| 3 | Region | ObjectDType | 0 (0.0%) | 1 (0.3%) | |||||
| 4 | Island | ObjectDType | 0 (0.0%) | 3 (0.9%) | |||||
| 5 | Stage | ObjectDType | 0 (0.0%) | 1 (0.3%) | |||||
| 6 | Individual ID | ObjectDType | 0 (0.0%) | 190 (55.2%) | |||||
| 7 | Clutch Completion | ObjectDType | 0 (0.0%) | 2 (0.6%) | |||||
| 8 | Date Egg | ObjectDType | 0 (0.0%) | 50 (14.5%) | |||||
| 9 | Culmen Length (mm) | Float64DType | 2 (0.6%) | 164 (47.7%) | 43.9 | 5.46 | 32.1 | 44.4 | 59.6 |
| 10 | Culmen Depth (mm) | Float64DType | 2 (0.6%) | 80 (23.3%) | 17.2 | 1.97 | 13.1 | 17.3 | 21.5 |
| 11 | Flipper Length (mm) | Float64DType | 2 (0.6%) | 55 (16.0%) | 201. | 14.1 | 172. | 197. | 231. |
| 12 | Body Mass (g) | Float64DType | 2 (0.6%) | 94 (27.3%) | 4.20e+03 | 802. | 2.70e+03 | 4.05e+03 | 6.30e+03 |
| 13 | Sex | ObjectDType | 10 (2.9%) | 3 (0.9%) | |||||
| 14 | Delta 15 N (o/oo) | Float64DType | 14 (4.1%) | 330 (95.9%) | 8.73 | 0.552 | 7.63 | 8.65 | 10.0 |
| 15 | Delta 13 C (o/oo) | Float64DType | 13 (3.8%) | 331 (96.2%) | -25.7 | 0.794 | -27.0 | -25.8 | -23.8 |
| 16 | Comments | ObjectDType | 290 (84.3%) | 10 (2.9%) |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
studyName
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Sample Number
Int64DType- Null values
- 0 (0.0%)
- Unique values
- 152 (44.2%)
- Mean ± Std
- 63.2 ± 40.4
- Median ± IQR
- 58 ± 66
- Min | Max
- 1 | 152
Species
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Region
ObjectDType- Null values
- 0 (0.0%)
Island
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Stage
ObjectDType- Null values
- 0 (0.0%)
Individual ID
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 190 (55.2%)
Most frequent values
Clutch Completion
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (0.6%)
Most frequent values
Date Egg
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 50 (14.5%)
Most frequent values
Culmen Length (mm)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 164 (47.7%)
- Mean ± Std
- 43.9 ± 5.46
- Median ± IQR
- 44.4 ± 9.30
- Min | Max
- 32.1 | 59.6
Culmen Depth (mm)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 80 (23.3%)
- Mean ± Std
- 17.2 ± 1.97
- Median ± IQR
- 17.3 ± 3.10
- Min | Max
- 13.1 | 21.5
Flipper Length (mm)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 55 (16.0%)
- Mean ± Std
- 201. ± 14.1
- Median ± IQR
- 197. ± 23.0
- Min | Max
- 172. | 231.
Body Mass (g)
Float64DType- Null values
- 2 (0.6%)
- Unique values
- 94 (27.3%)
- Mean ± Std
- 4.20e+03 ± 802.
- Median ± IQR
- 4.05e+03 ± 1.20e+03
- Min | Max
- 2.70e+03 | 6.30e+03
Sex
ObjectDType- Null values
- 10 (2.9%)
- Unique values
- 3 (0.9%)
Most frequent values
Delta 15 N (o/oo)
Float64DType- Null values
- 14 (4.1%)
- Unique values
- 330 (95.9%)
- Mean ± Std
- 8.73 ± 0.552
- Median ± IQR
- 8.65 ± 0.879
- Min | Max
- 7.63 | 10.0
Delta 13 C (o/oo)
Float64DType- Null values
- 13 (3.8%)
- Unique values
- 331 (96.2%)
- Mean ± Std
- -25.7 ± 0.794
- Median ± IQR
- -25.8 ± 1.27
- Min | Max
- -27.0 | -23.8
Comments
ObjectDType- Null values
- 290 (84.3%)
- Unique values
- 10 (2.9%)
Most frequent values
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| Clutch Completion | Comments | 0.992 |
| Delta 13 C (o/oo) | Comments | 0.818 |
| studyName | Sample Number | 0.781 |
| Flipper Length (mm) | Body Mass (g) | 0.757 |
| Culmen Depth (mm) | Flipper Length (mm) | 0.741 |
| Delta 15 N (o/oo) | Delta 13 C (o/oo) | 0.692 |
| Culmen Length (mm) | Flipper Length (mm) | 0.670 |
| Species | Flipper Length (mm) | 0.669 |
| Species | Island | 0.660 |
| Culmen Depth (mm) | Body Mass (g) | 0.651 |
| Sex | Comments | 0.629 |
| Flipper Length (mm) | Comments | 0.626 |
| Species | Body Mass (g) | 0.612 |
| Species | Culmen Depth (mm) | 0.605 |
| studyName | Date Egg | 0.599 |
| Delta 15 N (o/oo) | Comments | 0.588 |
| Species | Culmen Length (mm) | 0.579 |
| Culmen Length (mm) | Culmen Depth (mm) | 0.559 |
| Culmen Length (mm) | Body Mass (g) | 0.543 |
| Sample Number | Island | 0.525 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
We select features to predict penguin body mass. We remove rows with missing target values.
features = [
"Island",
"Clutch Completion",
"Flipper Length (mm)",
"Culmen Length (mm)",
"Culmen Depth (mm)",
"Species",
"Sex",
]
target = "Body Mass (g)"
data, target = penguins[features], penguins[target]
target = target.dropna()
data = data.loc[target.index]
data
Processing column 1 / 7
Processing column 2 / 7
Processing column 3 / 7
Processing column 4 / 7
Processing column 5 / 7
Processing column 6 / 7
Processing column 7 / 7
| Island | Clutch Completion | Flipper Length (mm) | Culmen Length (mm) | Culmen Depth (mm) | Species | Sex | |
|---|---|---|---|---|---|---|---|
| 0 | Torgersen | Yes | 181.0 | 39.1 | 18.7 | Adelie Penguin (Pygoscelis adeliae) | MALE |
| 1 | Torgersen | Yes | 186.0 | 39.5 | 17.4 | Adelie Penguin (Pygoscelis adeliae) | FEMALE |
| 2 | Torgersen | Yes | 195.0 | 40.3 | 18.0 | Adelie Penguin (Pygoscelis adeliae) | FEMALE |
| 4 | Torgersen | Yes | 193.0 | 36.7 | 19.3 | Adelie Penguin (Pygoscelis adeliae) | FEMALE |
| 5 | Torgersen | Yes | 190.0 | 39.3 | 20.6 | Adelie Penguin (Pygoscelis adeliae) | MALE |
| 339 | Dream | Yes | 207.0 | 55.8 | 19.8 | Chinstrap penguin (Pygoscelis antarctica) | MALE |
| 340 | Dream | No | 202.0 | 43.5 | 18.1 | Chinstrap penguin (Pygoscelis antarctica) | FEMALE |
| 341 | Dream | No | 193.0 | 49.6 | 18.2 | Chinstrap penguin (Pygoscelis antarctica) | MALE |
| 342 | Dream | Yes | 210.0 | 50.8 | 19.0 | Chinstrap penguin (Pygoscelis antarctica) | MALE |
| 343 | Dream | Yes | 198.0 | 50.2 | 18.7 | Chinstrap penguin (Pygoscelis antarctica) | FEMALE |
Island
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Clutch Completion
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (0.6%)
Most frequent values
Flipper Length (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 55 (16.1%)
- Mean ± Std
- 201. ± 14.1
- Median ± IQR
- 197. ± 23.0
- Min | Max
- 172. | 231.
Culmen Length (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 164 (48.0%)
- Mean ± Std
- 43.9 ± 5.46
- Median ± IQR
- 44.4 ± 9.30
- Min | Max
- 32.1 | 59.6
Culmen Depth (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 80 (23.4%)
- Mean ± Std
- 17.2 ± 1.97
- Median ± IQR
- 17.3 ± 3.10
- Min | Max
- 13.1 | 21.5
Species
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Sex
ObjectDType- Null values
- 8 (2.3%)
- Unique values
- 3 (0.9%)
Most frequent values
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Island | ObjectDType | 0 (0.0%) | 3 (0.9%) | |||||
| 1 | Clutch Completion | ObjectDType | 0 (0.0%) | 2 (0.6%) | |||||
| 2 | Flipper Length (mm) | Float64DType | 0 (0.0%) | 55 (16.1%) | 201. | 14.1 | 172. | 197. | 231. |
| 3 | Culmen Length (mm) | Float64DType | 0 (0.0%) | 164 (48.0%) | 43.9 | 5.46 | 32.1 | 44.4 | 59.6 |
| 4 | Culmen Depth (mm) | Float64DType | 0 (0.0%) | 80 (23.4%) | 17.2 | 1.97 | 13.1 | 17.3 | 21.5 |
| 5 | Species | ObjectDType | 0 (0.0%) | 3 (0.9%) | |||||
| 6 | Sex | ObjectDType | 8 (2.3%) | 3 (0.9%) |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
Island
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Clutch Completion
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (0.6%)
Most frequent values
Flipper Length (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 55 (16.1%)
- Mean ± Std
- 201. ± 14.1
- Median ± IQR
- 197. ± 23.0
- Min | Max
- 172. | 231.
Culmen Length (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 164 (48.0%)
- Mean ± Std
- 43.9 ± 5.46
- Median ± IQR
- 44.4 ± 9.30
- Min | Max
- 32.1 | 59.6
Culmen Depth (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 80 (23.4%)
- Mean ± Std
- 17.2 ± 1.97
- Median ± IQR
- 17.3 ± 3.10
- Min | Max
- 13.1 | 21.5
Species
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 3 (0.9%)
Most frequent values
Sex
ObjectDType- Null values
- 8 (2.3%)
- Unique values
- 3 (0.9%)
Most frequent values
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| Flipper Length (mm) | Species | 0.707 |
| Culmen Length (mm) | Species | 0.667 |
| Island | Species | 0.658 |
| Culmen Depth (mm) | Species | 0.639 |
| Island | Flipper Length (mm) | 0.512 |
| Island | Culmen Depth (mm) | 0.498 |
| Flipper Length (mm) | Culmen Depth (mm) | 0.387 |
| Island | Culmen Length (mm) | 0.351 |
| Culmen Depth (mm) | Sex | 0.339 |
| Flipper Length (mm) | Culmen Length (mm) | 0.333 |
| Culmen Length (mm) | Sex | 0.302 |
| Culmen Length (mm) | Culmen Depth (mm) | 0.283 |
| Flipper Length (mm) | Sex | 0.274 |
| Clutch Completion | Culmen Depth (mm) | 0.258 |
| Clutch Completion | Flipper Length (mm) | 0.197 |
| Clutch Completion | Species | 0.178 |
| Island | Clutch Completion | 0.157 |
| Clutch Completion | Culmen Length (mm) | 0.133 |
| Island | Sex | 0.117 |
| Species | Sex | 0.0774 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
Let’s evaluate a simple linear model using skrub’s preprocessing.
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import KFold, cross_validate
model = skrub.tabular_learner(estimator=LinearRegression())
model.set_output(transform="pandas")
cv = KFold(n_splits=5, shuffle=True, random_state=42)
cv_results = cross_validate(
model, data, target, cv=cv, return_estimator=True, return_train_score=True
)
pd.DataFrame(cv_results)[["train_score", "test_score"]]
Processing column 1 / 2
Processing column 2 / 2
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
| train_score | test_score | |
|---|---|---|
| 0 | 0.8822775776060422 | 0.8114847854917466 |
| 1 | 0.8795845281201589 | 0.8394348553838036 |
| 2 | 0.8697885501596277 | 0.8770481181740108 |
| 3 | 0.867805030683953 | 0.8868488072574447 |
| 4 | 0.8720016588218696 | 0.8725424597803983 |
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00631
- Median ± IQR
- 0.872 ± 0.00980
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.857 ± 0.0313
- Median ± IQR
- 0.873 ± 0.0376
- Min | Max
- 0.811 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | train_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.874 | 0.00631 | 0.868 | 0.872 | 0.882 |
| 1 | test_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.857 | 0.0313 | 0.811 | 0.873 | 0.887 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00631
- Median ± IQR
- 0.872 ± 0.00980
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.857 ± 0.0313
- Median ± IQR
- 0.873 ± 0.0376
- Min | Max
- 0.811 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| train_score | test_score | 1.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
The test score looks good overall but performs poorly on one fold. Let’s examine the coefficient values to understand why.
coefs = [est[-1].coef_ for est in cv_results["estimator"]]
coefs = pd.DataFrame(coefs, columns=cv_results["estimator"][0][-1].feature_names_in_)
coefs.plot.box(whis=[0, 100], vert=False)
plt.show()
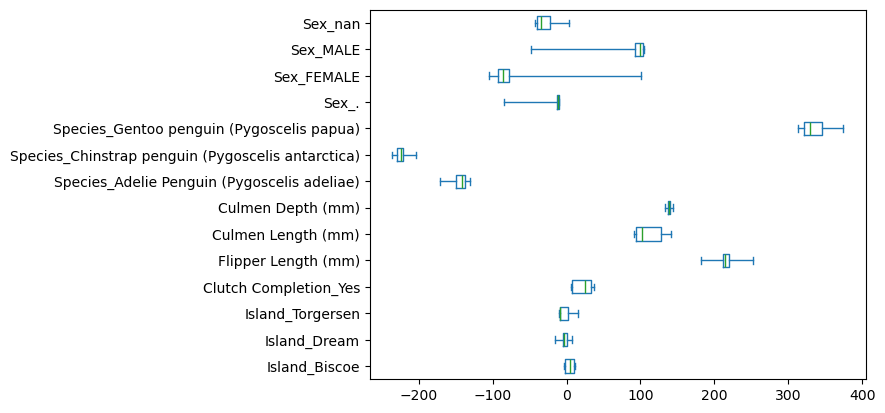
EXERCISE
What do you observe? What causes this behavior? Apply the preprocessing chain and check skrub’s statistics on the resulting data to understand these coefficients.
# Write your code here.
Ridge regressor - L2 regularization#
We saw that coefficients can grow arbitrarily large when features correlate.
L2 regularization forces weights toward zero. The parameter \(\alpha\) controls this shrinkage. Scikit-learn implements this as the Ridge model. Let’s fit it and examine its effect on weights.
from sklearn.linear_model import Ridge
model = skrub.tabular_learner(estimator=Ridge(alpha=1)).set_output(transform="pandas")
cv_results = cross_validate(
model, data, target, cv=cv, return_estimator=True, return_train_score=True
)
pd.DataFrame(cv_results)[["train_score", "test_score"]]
Processing column 1 / 2
Processing column 2 / 2
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
| train_score | test_score | |
|---|---|---|
| 0 | 0.8822424247171067 | 0.8122357028978447 |
| 1 | 0.8795604661243841 | 0.8393711829664781 |
| 2 | 0.8697645458439044 | 0.8767961310388296 |
| 3 | 0.8677758230915444 | 0.8868640133914093 |
| 4 | 0.8719740572383515 | 0.8726581033873201 |
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00631
- Median ± IQR
- 0.872 ± 0.00980
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.858 ± 0.0310
- Median ± IQR
- 0.873 ± 0.0374
- Min | Max
- 0.812 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | train_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.874 | 0.00631 | 0.868 | 0.872 | 0.882 |
| 1 | test_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.858 | 0.0310 | 0.812 | 0.873 | 0.887 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00631
- Median ± IQR
- 0.872 ± 0.00980
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.858 ± 0.0310
- Median ± IQR
- 0.873 ± 0.0374
- Min | Max
- 0.812 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| train_score | test_score | 1.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
coefs = [est[-1].coef_ for est in cv_results["estimator"]]
coefs = pd.DataFrame(coefs, columns=cv_results["estimator"][0][-1].feature_names_in_)
coefs.plot.box(whis=[0, 100], vert=False)
plt.show()
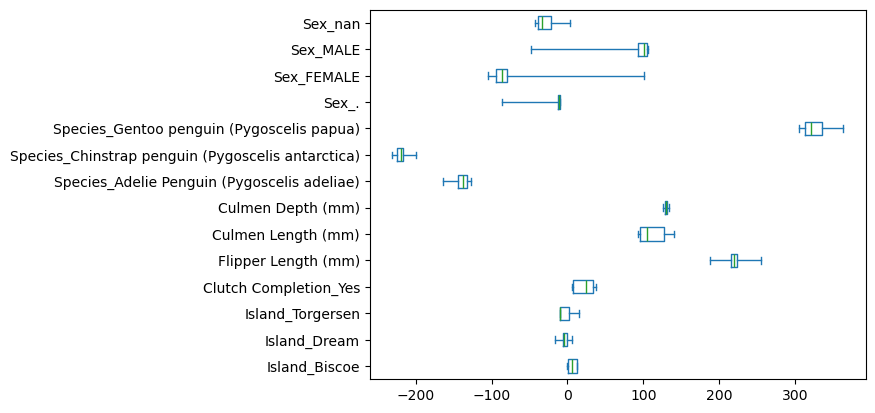
A small regularization solves the weight problem. We recover the original relationship:
EXERCISE
Try different \(\alpha\) values and examine how they affect the weights.
# Write your code here.
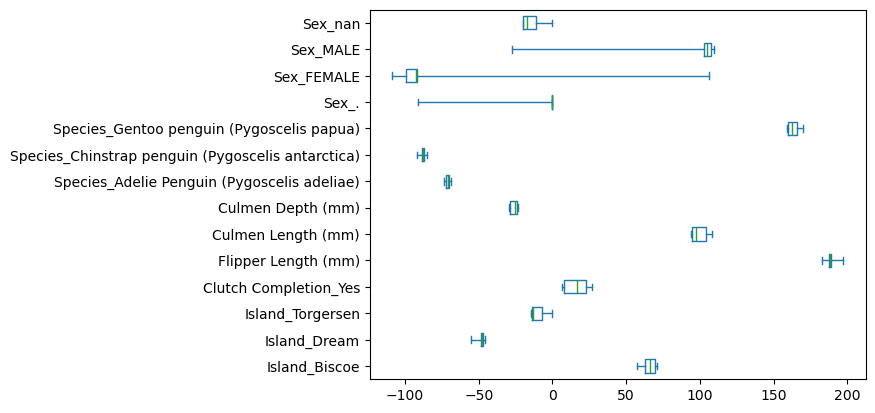
Lasso regressor - L1 regularization#
L1 provides another regularization type. It follows this formula:
Scikit-learn implements this as the Lasso regressor.
EXERCISE
Repeat the previous experiment with different \(\alpha\) values and examine how they affect the weights \(\beta\).
# Write your code here.
Elastic net - Combining L2 and L1 regularization#
Combining L2 and L1 regularization offers unique benefits: it identifies important features while preventing non-zero coefficients from growing too large.
from sklearn.linear_model import ElasticNet
model = skrub.tabular_learner(estimator=ElasticNet(alpha=10, l1_ratio=0.95))
model.set_output(transform="pandas")
cv_results = cross_validate(
model, data, target, cv=cv, return_estimator=True, return_train_score=True
)
pd.DataFrame(cv_results)[["train_score", "test_score"]]
Processing column 1 / 2
Processing column 2 / 2
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
| train_score | test_score | |
|---|---|---|
| 0 | 0.8483319045988991 | 0.7946495915893026 |
| 1 | 0.8465111734233646 | 0.816253456517587 |
| 2 | 0.8345425957172884 | 0.8456051730644178 |
| 3 | 0.8323476052428443 | 0.8577298889498699 |
| 4 | 0.8399405646701691 | 0.8355879567264349 |
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.840 ± 0.00706
- Median ± IQR
- 0.840 ± 0.0120
- Min | Max
- 0.832 | 0.848
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.830 ± 0.0249
- Median ± IQR
- 0.836 ± 0.0294
- Min | Max
- 0.795 | 0.858
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | train_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.840 | 0.00706 | 0.832 | 0.840 | 0.848 |
| 1 | test_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.830 | 0.0249 | 0.795 | 0.836 | 0.858 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.840 ± 0.00706
- Median ± IQR
- 0.840 ± 0.0120
- Min | Max
- 0.832 | 0.848
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.830 ± 0.0249
- Median ± IQR
- 0.836 ± 0.0294
- Min | Max
- 0.795 | 0.858
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| train_score | test_score | 1.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
coefs = [est[-1].coef_ for est in cv_results["estimator"]]
coefs = pd.DataFrame(coefs, columns=cv_results["estimator"][0][-1].feature_names_in_)
coefs.plot.box(whis=[0, 100], vert=False)
plt.show()
Hyperparameter tuning#
How do we choose the regularization parameter? The validation curve helps analyze single parameter effects. It plots scores versus parameter values.
Let’s use ValidationCurveDisplay to analyze how the alpha parameter affects Ridge regression.
model = skrub.tabular_learner(estimator=Ridge()).set_output(transform="pandas")
We need to find the parameter name for alpha in the model.
model.get_params()
{'memory': None,
'steps': [('tablevectorizer', TableVectorizer()),
('simpleimputer', SimpleImputer(add_indicator=True)),
('standardscaler', StandardScaler()),
('ridge', Ridge())],
'transform_input': None,
'verbose': False,
'tablevectorizer': TableVectorizer(),
'simpleimputer': SimpleImputer(add_indicator=True),
'standardscaler': StandardScaler(),
'ridge': Ridge(),
'tablevectorizer__cardinality_threshold': 40,
'tablevectorizer__datetime__add_total_seconds': True,
'tablevectorizer__datetime__add_weekday': False,
'tablevectorizer__datetime__resolution': 'hour',
'tablevectorizer__datetime': DatetimeEncoder(),
'tablevectorizer__drop_null_fraction': 1.0,
'tablevectorizer__high_cardinality__add_words': False,
'tablevectorizer__high_cardinality__analyzer': 'char',
'tablevectorizer__high_cardinality__batch_size': 1024,
'tablevectorizer__high_cardinality__gamma_scale_prior': 1.0,
'tablevectorizer__high_cardinality__gamma_shape_prior': 1.1,
'tablevectorizer__high_cardinality__hashing': False,
'tablevectorizer__high_cardinality__hashing_n_features': 4096,
'tablevectorizer__high_cardinality__init': 'k-means++',
'tablevectorizer__high_cardinality__max_iter': 5,
'tablevectorizer__high_cardinality__max_iter_e_step': 1,
'tablevectorizer__high_cardinality__max_no_improvement': 5,
'tablevectorizer__high_cardinality__n_components': 30,
'tablevectorizer__high_cardinality__ngram_range': (2, 4),
'tablevectorizer__high_cardinality__random_state': None,
'tablevectorizer__high_cardinality__rescale_W': True,
'tablevectorizer__high_cardinality__rescale_rho': False,
'tablevectorizer__high_cardinality__rho': 0.95,
'tablevectorizer__high_cardinality__verbose': 0,
'tablevectorizer__high_cardinality': GapEncoder(n_components=30),
'tablevectorizer__low_cardinality__categories': 'auto',
'tablevectorizer__low_cardinality__drop': 'if_binary',
'tablevectorizer__low_cardinality__dtype': 'float32',
'tablevectorizer__low_cardinality__feature_name_combiner': 'concat',
'tablevectorizer__low_cardinality__handle_unknown': 'ignore',
'tablevectorizer__low_cardinality__max_categories': None,
'tablevectorizer__low_cardinality__min_frequency': None,
'tablevectorizer__low_cardinality__sparse_output': False,
'tablevectorizer__low_cardinality': OneHotEncoder(drop='if_binary', dtype='float32', handle_unknown='ignore',
sparse_output=False),
'tablevectorizer__n_jobs': None,
'tablevectorizer__numeric': PassThrough(),
'tablevectorizer__specific_transformers': (),
'simpleimputer__add_indicator': True,
'simpleimputer__copy': True,
'simpleimputer__fill_value': None,
'simpleimputer__keep_empty_features': False,
'simpleimputer__missing_values': nan,
'simpleimputer__strategy': 'mean',
'standardscaler__copy': True,
'standardscaler__with_mean': True,
'standardscaler__with_std': True,
'ridge__alpha': 1.0,
'ridge__copy_X': True,
'ridge__fit_intercept': True,
'ridge__max_iter': None,
'ridge__positive': False,
'ridge__random_state': None,
'ridge__solver': 'auto',
'ridge__tol': 0.0001}
import numpy as np
from sklearn.model_selection import ValidationCurveDisplay
disp = ValidationCurveDisplay.from_estimator(
model,
data,
target,
cv=cv,
std_display_style="errorbar",
param_name="ridge__alpha",
param_range=np.logspace(-3, 3, num=20),
n_jobs=2,
)
plt.show()
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/linear_model/_ridge.py:215: LinAlgWarning: Ill-conditioned matrix (rcond=5.26528e-08): result may not be accurate.
return linalg.solve(A, Xy, assume_a="pos", overwrite_a=True).T
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
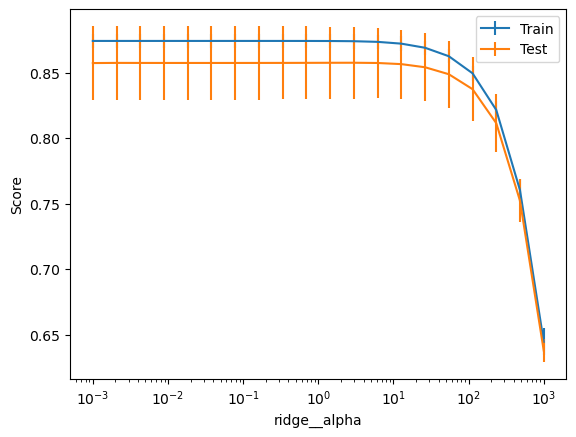
Too much regularization degrades model performance.
EXERCISE
Try a very small alpha (e.g. 1e-16) and observe its effect on the
validation curve.
# Write your code here.
In practice, we often use grid or random search instead of validation curves to choose regularization parameters. These methods run internal cross-validation to select the best-performing model. Let’s demonstrate random search.
from sklearn.model_selection import train_test_split
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=42
)
from scipy.stats import loguniform
from sklearn.model_selection import RandomizedSearchCV
param_distributions = {"ridge__alpha": loguniform(1e-3, 1e3)}
search = RandomizedSearchCV(model, param_distributions, n_iter=10, cv=cv)
search.fit(data_train, target_train)
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
RandomizedSearchCV(cv=KFold(n_splits=5, random_state=42, shuffle=True),
estimator=Pipeline(steps=[('tablevectorizer',
TableVectorizer()),
('simpleimputer',
SimpleImputer(add_indicator=True)),
('standardscaler',
StandardScaler()),
('ridge', Ridge())]),
param_distributions={'ridge__alpha': <scipy.stats._distn_infrastructure.rv_continuous_frozen object at 0x7fcefc77d280>})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomizedSearchCV(cv=KFold(n_splits=5, random_state=42, shuffle=True),
estimator=Pipeline(steps=[('tablevectorizer',
TableVectorizer()),
('simpleimputer',
SimpleImputer(add_indicator=True)),
('standardscaler',
StandardScaler()),
('ridge', Ridge())]),
param_distributions={'ridge__alpha': <scipy.stats._distn_infrastructure.rv_continuous_frozen object at 0x7fcefc77d280>})Pipeline(steps=[('tablevectorizer', TableVectorizer()),
('simpleimputer', SimpleImputer(add_indicator=True)),
('standardscaler', StandardScaler()),
('ridge', Ridge(alpha=np.float64(1.984293246098125)))])TableVectorizer()
['Flipper Length (mm)', 'Culmen Length (mm)', 'Culmen Depth (mm)']
PassThrough()
[]
DatetimeEncoder()
['Island', 'Clutch Completion', 'Species', 'Sex']
OneHotEncoder(drop='if_binary', dtype='float32', handle_unknown='ignore',
sparse_output=False)[]
GapEncoder(n_components=30)
SimpleImputer(add_indicator=True)
StandardScaler()
Ridge(alpha=np.float64(1.984293246098125))
search.best_params_
{'ridge__alpha': np.float64(1.984293246098125)}
pd.DataFrame(search.cv_results_)
Processing column 1 / 14
Processing column 2 / 14
Processing column 3 / 14
Processing column 4 / 14
Processing column 5 / 14
Processing column 6 / 14
Processing column 7 / 14
Processing column 8 / 14
Processing column 9 / 14
Processing column 10 / 14
Processing column 11 / 14
Processing column 12 / 14
Processing column 13 / 14
Processing column 14 / 14
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_ridge__alpha | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.035968017578125 | 0.001266300723086415 | 0.020697402954101562 | 0.0005012212587209833 | 234.458146774326 | {'ridge__alpha': np.float64(234.458146774326)} | 0.7748330689656573 | 0.8012270634196167 | 0.7976215582849489 | 0.8440499641656398 | 0.8217874564879523 | 0.8079038222647629 | 0.02342339710015936 | 9 |
| 1 | 0.03455343246459961 | 0.0006928242997513938 | 0.020396089553833006 | 0.00027681814366081346 | 19.83922736446405 | {'ridge__alpha': np.float64(19.83922736446405)} | 0.8287822466607332 | 0.8673629058812413 | 0.8730339048090712 | 0.8845213623446101 | 0.8808455371286077 | 0.8669091913648528 | 0.01997894984214285 | 7 |
| 2 | 0.03432607650756836 | 0.00019603285345577037 | 0.020389604568481445 | 0.0001498520534328059 | 0.09552649386031474 | {'ridge__alpha': np.float64(0.09552649386031474)} | 0.8383298420092085 | 0.8739740480744762 | 0.8871174251186338 | 0.8795669446687323 | 0.8756965092542 | 0.8709369538250501 | 0.01691972484160644 | 4 |
| 3 | 0.03453502655029297 | 0.00020953467249328097 | 0.020430994033813477 | 0.0002139492078187129 | 53.312147000652246 | {'ridge__alpha': np.float64(53.312147000652246)} | 0.81927678454271 | 0.8570145020610281 | 0.8592017925804524 | 0.8836753260060273 | 0.8725527378471669 | 0.858344228607477 | 0.02178217163020697 | 8 |
| 4 | 0.03485980033874512 | 0.0009625789555861882 | 0.020426559448242187 | 0.0003895558628703161 | 0.15932706110628528 | {'ridge__alpha': np.float64(0.15932706110628528)} | 0.8382944489903339 | 0.8739673815348152 | 0.8870484470733806 | 0.8796190100124445 | 0.8758384749640349 | 0.8709535525150018 | 0.01693338985681266 | 3 |
| 5 | 0.0342012882232666 | 0.00033350106747180147 | 0.02018399238586426 | 0.0002151431660354646 | 1.984293246098125 | {'ridge__alpha': np.float64(1.984293246098125)} | 0.8372737318652337 | 0.8736076787806398 | 0.8851701249776798 | 0.8808444289250773 | 0.8788742575609905 | 0.8711540444219242 | 0.017342731373012163 | 1 |
| 6 | 0.03408665657043457 | 0.0003194996629014024 | 0.02039036750793457 | 0.00045744628135241947 | 0.08883763971045147 | {'ridge__alpha': np.float64(0.08883763971045147)} | 0.8383333389266531 | 0.8739746987394621 | 0.8871246894226563 | 0.8795614317097146 | 0.8756815996531858 | 0.8709351516903343 | 0.016918390283742035 | 5 |
| 7 | 0.03484287261962891 | 0.0010020964669962395 | 0.02093191146850586 | 0.0009978201151210674 | 13.810982638149506 | {'ridge__alpha': np.float64(13.810982638149506)} | 0.8311879256489517 | 0.8694500817260187 | 0.8763243721726487 | 0.883981395653028 | 0.8819120910594156 | 0.8685711732520126 | 0.01935896080554444 | 6 |
| 8 | 0.0343986988067627 | 0.0002716848325260403 | 0.020312929153442384 | 0.00014484933143290847 | 2.4868137972895186 | {'ridge__alpha': np.float64(2.4868137972895186)} | 0.8369839325331889 | 0.8734687911277235 | 0.8846847742613769 | 0.8811115302581726 | 0.8794449202760392 | 0.8711387896913003 | 0.017458088279432855 | 2 |
| 9 | 0.03456645011901856 | 0.0006608197895915777 | 0.0204984188079834 | 0.00017287715223465743 | 762.6496480178704 | {'ridge__alpha': np.float64(762.6496480178704)} | 0.6295466008618905 | 0.6418659766719412 | 0.6320775726967458 | 0.6844970865880557 | 0.666179630075751 | 0.6508333733788769 | 0.0212324003765637 | 10 |
mean_fit_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.0346 ± 0.000530
- Median ± IQR
- 0.0345 ± 0.000517
- Min | Max
- 0.0341 | 0.0360
std_fit_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.000591 ± 0.000383
- Median ± IQR
- 0.000334 ± 0.000691
- Min | Max
- 0.000196 | 0.00127
mean_score_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.0205 ± 0.000209
- Median ± IQR
- 0.0204 ± 0.000109
- Min | Max
- 0.0202 | 0.0209
std_score_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.000352 ± 0.000261
- Median ± IQR
- 0.000215 ± 0.000285
- Min | Max
- 0.000145 | 0.000998
param_ridge__alpha
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 109. ± 241.
- Median ± IQR
- 2.49 ± 53.2
- Min | Max
- 0.0888 | 763.
params
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
Most frequent values
split0_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.807 ± 0.0654
- Median ± IQR
- 0.831 ± 0.0190
- Min | Max
- 0.630 | 0.838
split1_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.841 ± 0.0733
- Median ± IQR
- 0.869 ± 0.0170
- Min | Max
- 0.642 | 0.874
split2_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.847 ± 0.0803
- Median ± IQR
- 0.876 ± 0.0278
- Min | Max
- 0.632 | 0.887
split3_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.858 ± 0.0622
- Median ± IQR
- 0.880 ± 0.00411
- Min | Max
- 0.684 | 0.885
split4_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.851 ± 0.0673
- Median ± IQR
- 0.876 ± 0.00689
- Min | Max
- 0.666 | 0.882
mean_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.841 ± 0.0695
- Median ± IQR
- 0.869 ± 0.0126
- Min | Max
- 0.651 | 0.871
std_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.0191 ± 0.00239
- Median ± IQR
- 0.0175 ± 0.00430
- Min | Max
- 0.0169 | 0.0234
rank_test_score
Int32DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 5.50 ± 3.03
- Median ± IQR
- 5 ± 5
- Min | Max
- 1 | 10
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | mean_fit_time | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.0346 | 0.000530 | 0.0341 | 0.0345 | 0.0360 |
| 1 | std_fit_time | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.000591 | 0.000383 | 0.000196 | 0.000334 | 0.00127 |
| 2 | mean_score_time | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.0205 | 0.000209 | 0.0202 | 0.0204 | 0.0209 |
| 3 | std_score_time | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.000352 | 0.000261 | 0.000145 | 0.000215 | 0.000998 |
| 4 | param_ridge__alpha | Float64DType | 0 (0.0%) | 10 (100.0%) | 109. | 241. | 0.0888 | 2.49 | 763. |
| 5 | params | ObjectDType | 0 (0.0%) | 10 (100.0%) | |||||
| 6 | split0_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.807 | 0.0654 | 0.630 | 0.831 | 0.838 |
| 7 | split1_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.841 | 0.0733 | 0.642 | 0.869 | 0.874 |
| 8 | split2_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.847 | 0.0803 | 0.632 | 0.876 | 0.887 |
| 9 | split3_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.858 | 0.0622 | 0.684 | 0.880 | 0.885 |
| 10 | split4_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.851 | 0.0673 | 0.666 | 0.876 | 0.882 |
| 11 | mean_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.841 | 0.0695 | 0.651 | 0.869 | 0.871 |
| 12 | std_test_score | Float64DType | 0 (0.0%) | 10 (100.0%) | 0.0191 | 0.00239 | 0.0169 | 0.0175 | 0.0234 |
| 13 | rank_test_score | Int32DType | 0 (0.0%) | 10 (100.0%) | 5.50 | 3.03 | 1 | 5 | 10 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
mean_fit_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.0346 ± 0.000530
- Median ± IQR
- 0.0345 ± 0.000517
- Min | Max
- 0.0341 | 0.0360
std_fit_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.000591 ± 0.000383
- Median ± IQR
- 0.000334 ± 0.000691
- Min | Max
- 0.000196 | 0.00127
mean_score_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.0205 ± 0.000209
- Median ± IQR
- 0.0204 ± 0.000109
- Min | Max
- 0.0202 | 0.0209
std_score_time
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.000352 ± 0.000261
- Median ± IQR
- 0.000215 ± 0.000285
- Min | Max
- 0.000145 | 0.000998
param_ridge__alpha
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 109. ± 241.
- Median ± IQR
- 2.49 ± 53.2
- Min | Max
- 0.0888 | 763.
params
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
Most frequent values
split0_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.807 ± 0.0654
- Median ± IQR
- 0.831 ± 0.0190
- Min | Max
- 0.630 | 0.838
split1_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.841 ± 0.0733
- Median ± IQR
- 0.869 ± 0.0170
- Min | Max
- 0.642 | 0.874
split2_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.847 ± 0.0803
- Median ± IQR
- 0.876 ± 0.0278
- Min | Max
- 0.632 | 0.887
split3_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.858 ± 0.0622
- Median ± IQR
- 0.880 ± 0.00411
- Min | Max
- 0.684 | 0.885
split4_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.851 ± 0.0673
- Median ± IQR
- 0.876 ± 0.00689
- Min | Max
- 0.666 | 0.882
mean_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.841 ± 0.0695
- Median ± IQR
- 0.869 ± 0.0126
- Min | Max
- 0.651 | 0.871
std_test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 0.0191 ± 0.00239
- Median ± IQR
- 0.0175 ± 0.00430
- Min | Max
- 0.0169 | 0.0234
rank_test_score
Int32DType- Null values
- 0 (0.0%)
- Unique values
- 10 (100.0%)
- Mean ± Std
- 5.50 ± 3.03
- Median ± IQR
- 5 ± 5
- Min | Max
- 1 | 10
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| std_test_score | rank_test_score | 1.00 |
| mean_test_score | std_test_score | 1.00 |
| split4_test_score | rank_test_score | 1.00 |
| split3_test_score | std_test_score | 1.00 |
| split4_test_score | std_test_score | 1.00 |
| split4_test_score | mean_test_score | 1.00 |
| split3_test_score | rank_test_score | 1.00 |
| split3_test_score | split4_test_score | 1.00 |
| split3_test_score | mean_test_score | 1.00 |
| split2_test_score | rank_test_score | 1.00 |
| split2_test_score | std_test_score | 1.00 |
| split1_test_score | split3_test_score | 1.00 |
| split2_test_score | mean_test_score | 1.00 |
| split2_test_score | split4_test_score | 1.00 |
| split2_test_score | split3_test_score | 1.00 |
| split1_test_score | rank_test_score | 1.00 |
| split1_test_score | std_test_score | 1.00 |
| split1_test_score | mean_test_score | 1.00 |
| split1_test_score | split4_test_score | 1.00 |
| split0_test_score | split3_test_score | 1.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
This approach enables nested cross-validation. The inner loop selects parameters while the outer loop evaluates model performance.
cv_results = cross_validate(
search, data, target, cv=cv, return_estimator=True, return_train_score=True
)
pd.DataFrame(cv_results)[["train_score", "test_score"]]
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
Processing column 1 / 2
Processing column 2 / 2
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
| train_score | test_score | |
|---|---|---|
| 0 | 0.8822775787631696 | 0.8114838667117166 |
| 1 | 0.8795790993895044 | 0.839408678442042 |
| 2 | 0.8696825746795493 | 0.876501283964341 |
| 3 | 0.8677924887841242 | 0.8868648592306861 |
| 4 | 0.8719901926066118 | 0.8726274555009443 |
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00633
- Median ± IQR
- 0.872 ± 0.00990
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.857 ± 0.0312
- Median ± IQR
- 0.873 ± 0.0371
- Min | Max
- 0.811 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | train_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.874 | 0.00633 | 0.868 | 0.872 | 0.882 |
| 1 | test_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.857 | 0.0312 | 0.811 | 0.873 | 0.887 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00633
- Median ± IQR
- 0.872 ± 0.00990
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.857 ± 0.0312
- Median ± IQR
- 0.873 ± 0.0371
- Min | Max
- 0.811 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| train_score | test_score | 1.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
Some scikit-learn models efficiently search hyperparameters internally. Models with “CV” in their name, like RidgeCV, automatically find optimal regularization parameters.
from sklearn.linear_model import RidgeCV
model = skrub.tabular_learner(estimator=RidgeCV(alphas=np.logspace(-3, 3, num=100)))
model.set_output(transform="pandas")
cv_results = cross_validate(
model, data, target, cv=cv, return_estimator=True, return_train_score=True
)
pd.DataFrame(cv_results)[["train_score", "test_score"]]
Processing column 1 / 2
Processing column 2 / 2
/home/runner/work/traces-sklearn/traces-sklearn/.pixi/envs/docs/lib/python3.12/site-packages/sklearn/preprocessing/_encoders.py:246: UserWarning: Found unknown categories in columns [0] during transform. These unknown categories will be encoded as all zeros
warnings.warn(
| train_score | test_score | |
|---|---|---|
| 0 | 0.8822096843692167 | 0.8125020253713926 |
| 1 | 0.8794538214318453 | 0.8392377548970351 |
| 2 | 0.8695733005132162 | 0.8762494607310561 |
| 3 | 0.8676484731701924 | 0.886807985984249 |
| 4 | 0.8718116496306133 | 0.8726517794922556 |
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00636
- Median ± IQR
- 0.872 ± 0.00988
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.857 ± 0.0308
- Median ± IQR
- 0.873 ± 0.0370
- Min | Max
- 0.813 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | train_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.874 | 0.00636 | 0.868 | 0.872 | 0.882 |
| 1 | test_score | Float64DType | 0 (0.0%) | 5 (100.0%) | 0.857 | 0.0308 | 0.813 | 0.873 | 0.887 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
train_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.874 ± 0.00636
- Median ± IQR
- 0.872 ± 0.00988
- Min | Max
- 0.868 | 0.882
test_score
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 0.857 ± 0.0308
- Median ± IQR
- 0.873 ± 0.0370
- Min | Max
- 0.813 | 0.887
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| train_score | test_score | 1.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
alphas = [est[-1].alpha_ for est in cv_results["estimator"]]
alphas
[np.float64(1.4174741629268048),
np.float64(2.4770763559917115),
np.float64(3.2745491628777286),
np.float64(2.4770763559917115),
np.float64(2.848035868435802)]
What about classification?#
Classification handles regularization differently. Instead of creating new estimators, regularization becomes a model parameter. LogisticRegression and LinearSVC offer two main models. Both use penalty and C parameters (C inverts regression’s alpha).
We’ll explore parameter C with LogisticRegression. First, let’s load classification data to predict penguin species from culmen measurements.
data = pd.read_csv("../datasets/penguins_classification.csv")
data = data[data["Species"].isin(["Adelie", "Chinstrap"])]
data["Species"] = data["Species"].astype("category")
data.head()
Processing column 1 / 3
Processing column 2 / 3
Processing column 3 / 3
| Culmen Length (mm) | Culmen Depth (mm) | Species | |
|---|---|---|---|
| 0 | 39.1 | 18.7 | Adelie |
| 1 | 39.5 | 17.4 | Adelie |
| 2 | 40.3 | 18.0 | Adelie |
| 3 | 36.7 | 19.3 | Adelie |
| 4 | 39.3 | 20.6 | Adelie |
Culmen Length (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 39.0 ± 1.35
- Median ± IQR
- 39.3 ± 0.400
- Min | Max
- 36.7 | 40.3
Culmen Depth (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 18.8 ± 1.23
- Median ± IQR
- 18.7 ± 1.30
- Min | Max
- 17.4 | 20.6
Species
CategoricalDtype- Null values
- 0 (0.0%)
- Unique values
- 2 (40.0%)
Most frequent values
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column | Column name | dtype | Null values | Unique values | Mean | Std | Min | Median | Max |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Culmen Length (mm) | Float64DType | 0 (0.0%) | 5 (100.0%) | 39.0 | 1.35 | 36.7 | 39.3 | 40.3 |
| 1 | Culmen Depth (mm) | Float64DType | 0 (0.0%) | 5 (100.0%) | 18.8 | 1.23 | 17.4 | 18.7 | 20.6 |
| 2 | Species | CategoricalDtype | 0 (0.0%) | 2 (40.0%) |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
Culmen Length (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 39.0 ± 1.35
- Median ± IQR
- 39.3 ± 0.400
- Min | Max
- 36.7 | 40.3
Culmen Depth (mm)
Float64DType- Null values
- 0 (0.0%)
- Unique values
- 5 (100.0%)
- Mean ± Std
- 18.8 ± 1.23
- Median ± IQR
- 18.7 ± 1.30
- Min | Max
- 17.4 | 20.6
Species
CategoricalDtype- Null values
- 0 (0.0%)
- Unique values
- 2 (40.0%)
Most frequent values
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V |
|---|---|---|
| Culmen Length (mm) | Culmen Depth (mm) | 1.00 |
| Culmen Depth (mm) | Species | 0.00 |
| Culmen Length (mm) | Species | 0.00 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
X, y = data[["Culmen Length (mm)", "Culmen Depth (mm)"]], data["Species"]
import matplotlib.pyplot as plt
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
edgecolor="black",
s=50,
)
plt.show()
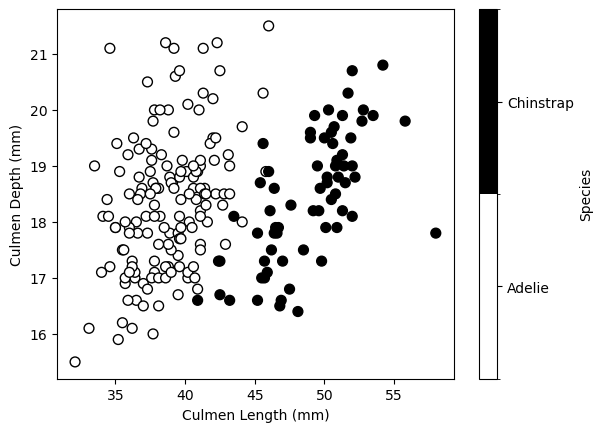
QUESTION
What regularization does LogisticRegression use by default? Check the documentation.
Let’s fit a model and visualize its decision boundary.
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X, y)
LogisticRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression()
from sklearn.inspection import DecisionBoundaryDisplay
display = DecisionBoundaryDisplay.from_estimator(
model,
X,
response_method="decision_function",
cmap=plt.cm.RdBu,
plot_method="pcolormesh",
shading="auto",
)
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
edgecolor="black",
s=50,
ax=display.ax_,
)
plt.show()
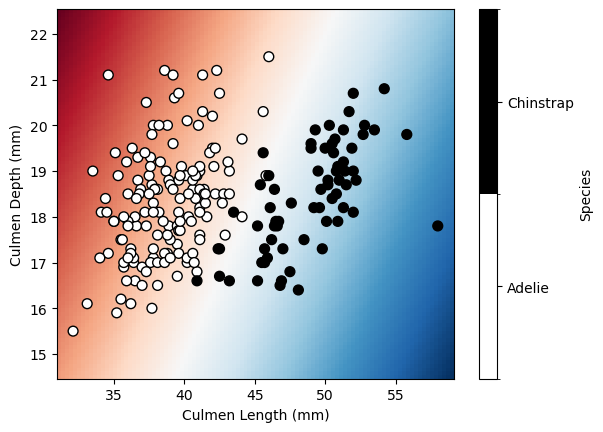
coef = pd.Series(model.coef_[0], index=X.columns)
coef.plot.barh()
plt.show()
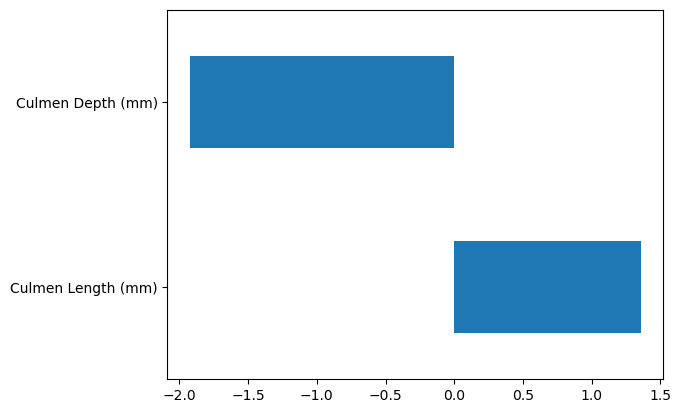
This example establishes a baseline for studying parameter C effects. The logistic regression loss function is:
EXERCISE
Fit models with different C values and examine how they affect coefficients and decision boundaries.
# Write your code here.
The loss formula shows C affects the data term (error between true and predicted targets). In regression, alpha affects the weights instead. This explains why C inversely relates to alpha.